MOLECULAR CHARACTERIZATION OF Aspergillus flavus TOXIGENICITY IN AGRICULTURAL COMMODITIES IN INDONESIA
Downloads
Toxigenic Aspergillus flavus is a primary producer of aflatoxin in Indonesia, and its presence can lead to the contamination of agricultural commodities. This contamination poses a risk to export-targeted commodities, potentially resulting in their rejection. Therefore, this study aims to characterize the molecular profile of native
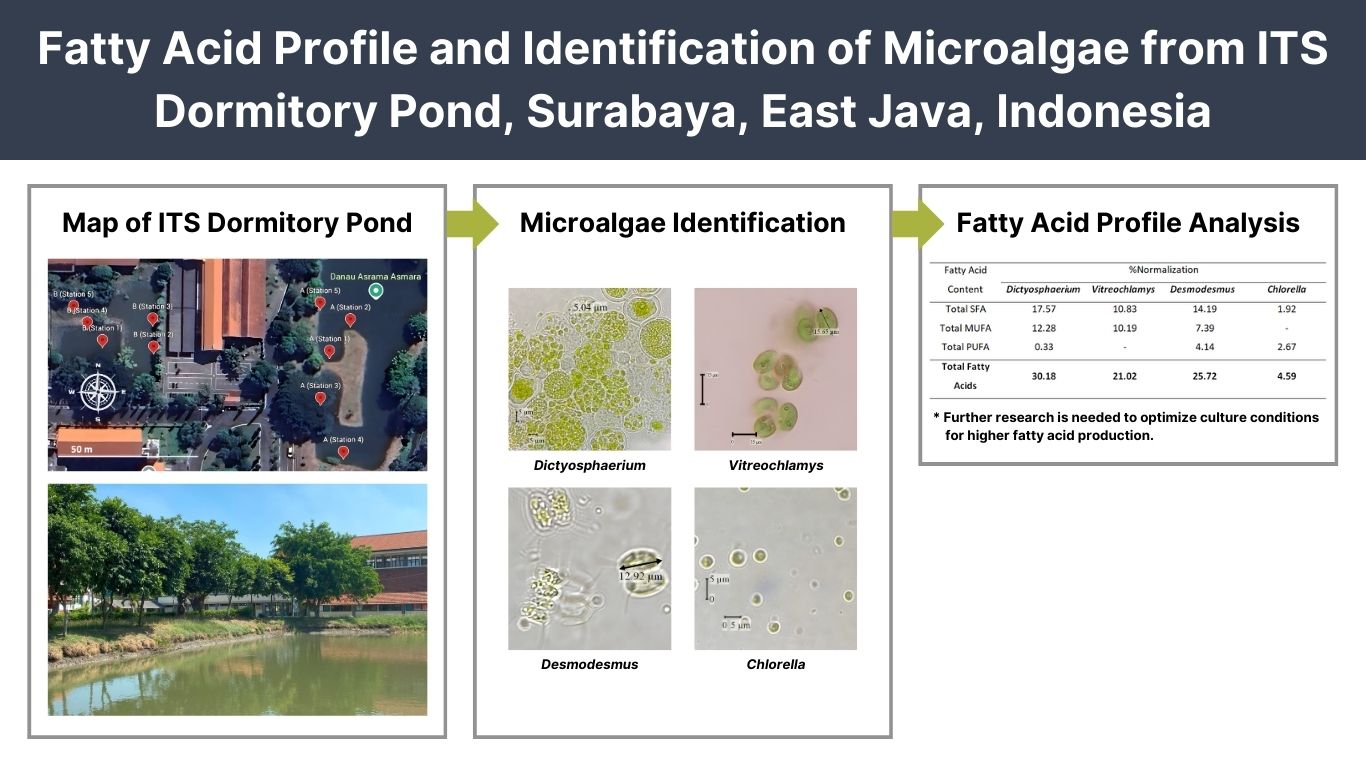
A. flavus isolated from several Indonesian agricultural products, with a major focus on its toxigenicity and toxin production. A total of 18 A. flavus collections were isolated from nutmeg, ground peanut, cacao, coffee bean, corn, white pepper, and soil peanut plantation. Species identification was carried out using molecular and morphological approaches. The toxigenicity of isolates was characterized based on the amplification of aflatoxin gene clusters, while toxin production was assessed through growth simulation on a 10% coconut broth media followed by HPLC quantification. The result showed that all isolates were confirmed as A. flavus based on the morphological and sequence analysis of the ITS region. A total of 11 isolates (61%) were confirmed as toxigenic and produced 1-2 types of aflatoxin, in varying concentrations of high, moderate, or low levels of AFB1. High levels of AFB1 produced by seven isolates namely BIO3313, BIO33212, BIO3361, BIO33404, BIO3338, BIO3352, and BIO3344, had concentration levels ranging from 76.78 to 2241.06 µg/kg, while three isolates (BIO3314, BIO3312, and BIO3381) produced AFB1 below 1 µg/kg. Twenty-nine pairs of aflatoxin gene-specific sequences were successfully amplified as a single band, while some produced non-specific patterns in several low toxigenic and non-toxigenic isolates. Based on the results, it was concluded that completed gene clusters and variations of gene deletion were observed in both toxigenic and non-toxigenic isolates. However, no specific target gene could effectively distinguish the two groups. Two non-toxigenic isolates namely BIO3393 and BIO33403 exhibited a large deletion and could be potential candidates for biocontrol agents.
Downloads
Anidah, Rahayu WP, Nurjanah S. 2020. Screening of toxigenic Aspergillus flavus strains and aflatoxin content from agricultural commodities in Indonesia. In: Sitanggang AZ, Rahayu WP, Antara NS, Santoso U, Giyatmi, Ardiansyah, Haryono A, Eds. Proceedings of the 16th ASEAN Food Conference-16th AFC, ISBN 978-989-758-467-1, p. 221-6. DOI: 10.5220/ 0009981202210226
AOAC. 2000. Section 49.2.18 (AOAC Method 991.31) for corn, raw peanut, and peanut butter. In Official method of analysis. 17th Edition. Gaithersburg (US): AOAC International.
Bhat R, Rai RV, Karim AA. 2010. Mycotoxins in food and feed: present status and future concerns. Compr Rev Food Sci Food Saf 9: 57-81.
[BPOM] Badan Pengawas Obat dan Makanan. 2018. Indonesian POM Agency Regulation No. 8 of 2018. The Maximum Limit of Chemical Contamination in Processed Food. Jakarta (ID): Badan Pengawas Obat dan Makanan.
Chang PK, Horn BW, Dorner JW. 2005. Sequence breakpoint in the aflatoxin biosynthesis gene cluster and flanking region in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet Biol 42(11): 914-23.
Criseo G, Bagnara A, Bisignano G. 2001. Differentiation of aflatoxin-producing and non-producing strains of Aspergillus flavus group. Appl Microbiol 33(4): 291-5.
Davari E, Mohsenzadeh M, Mohammadi Gh, Rezaeian-Doloei R. 2014. Characterization of aflatoxigenic Aspergillus flavus and A. paraciticus isolates from animal feedstuffs in northeastern Iran. IJVR 16(2): 150-5.
Davis ND, Iyer SK, Diener UL. 1987. Improved method of screening for aflatoxin with a coconut agar medium. Appl Environ Microbiol 53(7): 1593-5.
Degola F, Berni E, Dall’Asta C, Spotti E, Marchelli R, Ferrero I, Restivo FM. 2007. A multiplex RT-PCR approach to detect aflatoxigenic strains of Aspergillus flavus. J Appl Microbiol 103(2):409-17. DOI: 10.1111/j.1365-2672.2006.03256.x
Dharmaputra OS. 2002. Review on aflatoxin in Indonesian food and feedstuffs and their products. Biotropia 19: 26-46.
Donner M, Atehnkeng J, Sikora, RA, Bandyopadhyay R, Cotty PJ. 2010. Molecular characterization of a toxigenic strains for biological control of aflatoxins in Nigeria. Food Addit Contam 27(5): 576-90. DOI: 10.1080/19440040903551954
Kim DM, Chung SH, Chun HS. 2011. Multiplex PCR assay for the detection of aflatoxigenic and non-aflatoxigenic fungi in Meju, a Korean fermented soybean food starter. Food Microbiol 28: 1402-8.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547-9. DOI:10.1093/ molbev/msy096
Levin RE. 2012. PCR detection of aflatoxin producing fungi and its limitations. Int J Food Microbiol 156(1): 1-6.
Lin MT, Dianese JC. 1976. A coconut-agar medium for rapid detection of aflatoxin production by Aspergillus spp. Phytopathology 66: 1466-9.
Mamo FT, Selvaraj JN, Wang Y, Liu Y. 2017. Recent developments in the screening of atoxigenic Aspergillus flavus towards aflatoxin biocontrol. Appl Environ Microbiol 5(1): 20-30.
Pitt JI, Hocking AD. 2009. Fungi and food spoilage. 3rd Edition. New York (US): Springer. 519 p. Qiagen. 2019. DNeasy Plant Handbook. Germany.
Rao KR, Vipin AV, Venkateswaran G. 2020. Molecular profile of non-aflatoxigenic phenotype in native strains of Aspergillus flavus. Arch Microbiol 202(5):1143-55. DOI:10.1007/s00203-020-01822-1
Samson RA, Hoekstra ES, Frisvad JC. 2004. Introduction to food and airborne fungi. 7th Edition. Utrecht (NL): Centraalbureau voor Schimmelcultures (CBS).
Schmidt-Heydt M, Magan N, Geisen R. 2008. Stress induction of mycotoxin biosynthesis genes by abiotic factors. FEMS Microbiol Lett 84(2): 142-9. DOI:10.1111/j.1574-6968.2008.01182.x
Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) Software version 4.0. Mol Biol Evol 24(8): 1596-9.
Tran-Dinh N, Pitt JI, Markwell PJ. 2014 Selection of non-toxigenic strains of Aspergillus favus for biocontrol of afatoxins in maize in Thailand. Biocontrol Sci Technol 24: 652-66.
Wei D, Zhou L, Selvaraj JN, Zhang C, Xing F, Zhao Y, Wang Y, Liu Y. 2014. Molecular characterization of a toxigenic Aspergillus flavus isolates collected in China. J Microbiol 52:559-65. DOI:10.1007/ s12275-014-3629-8.
White TJ, Bruns TD, Lee SB, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, Eds. PCR Protocols. A guide to methods and applications. San Diego (US): Academic Press. p. 315-20.
Yin Y, Lou T, Yan L, Michailides TJ, Ma Z. 2009. Molecular characterization of toxigenic and a toxigenic Aspergillus flavus isolates, collected from peanut fields in China. J Appl Microbiol 107: 1857-65.
Yu J, Bhatnagar D, Ehrlich KC. 2002. Aflatoxin biosynthesis. Iberoam Micol 19: 191-200.
Copyright (c) 2023 ANIDAH, WINIATI P. RAHAYU, SITI NURJANAHAND INA RETNOWATI

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Authors who publish with this journal agree with the following terms:
- Authors retain copyright and grant the journal right of first publication, with the work 1 year after publication simultaneously licensed under a Creative Commons attribution-noncommerical-noderivates 4.0 International License that allows others to share, copy and redistribute the work in any medium or format, but only where the use is for non-commercial purposes and an acknowledgement of the work's authorship and initial publication in this journal is mentioned.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See The Effect of Open Access).