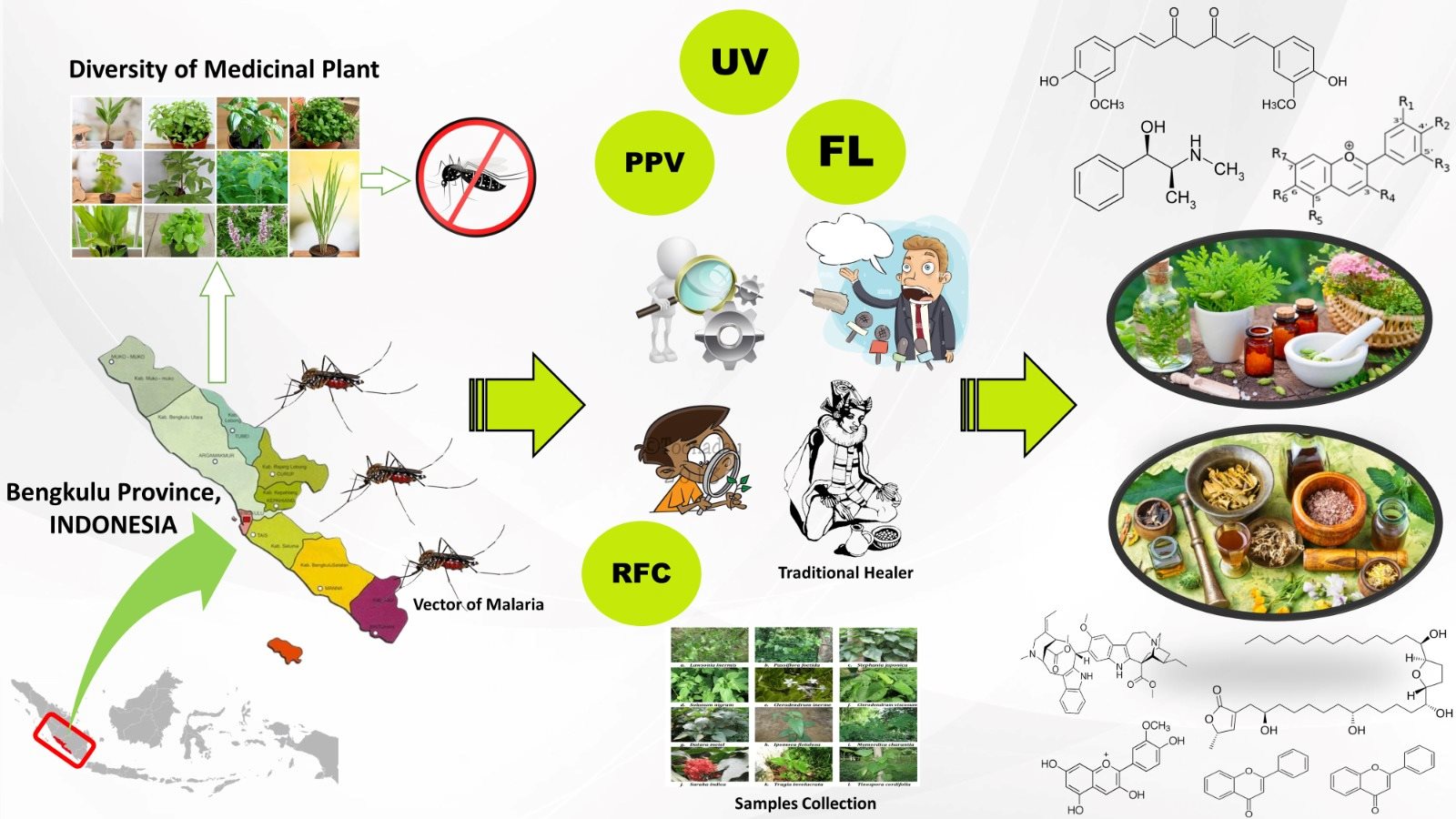
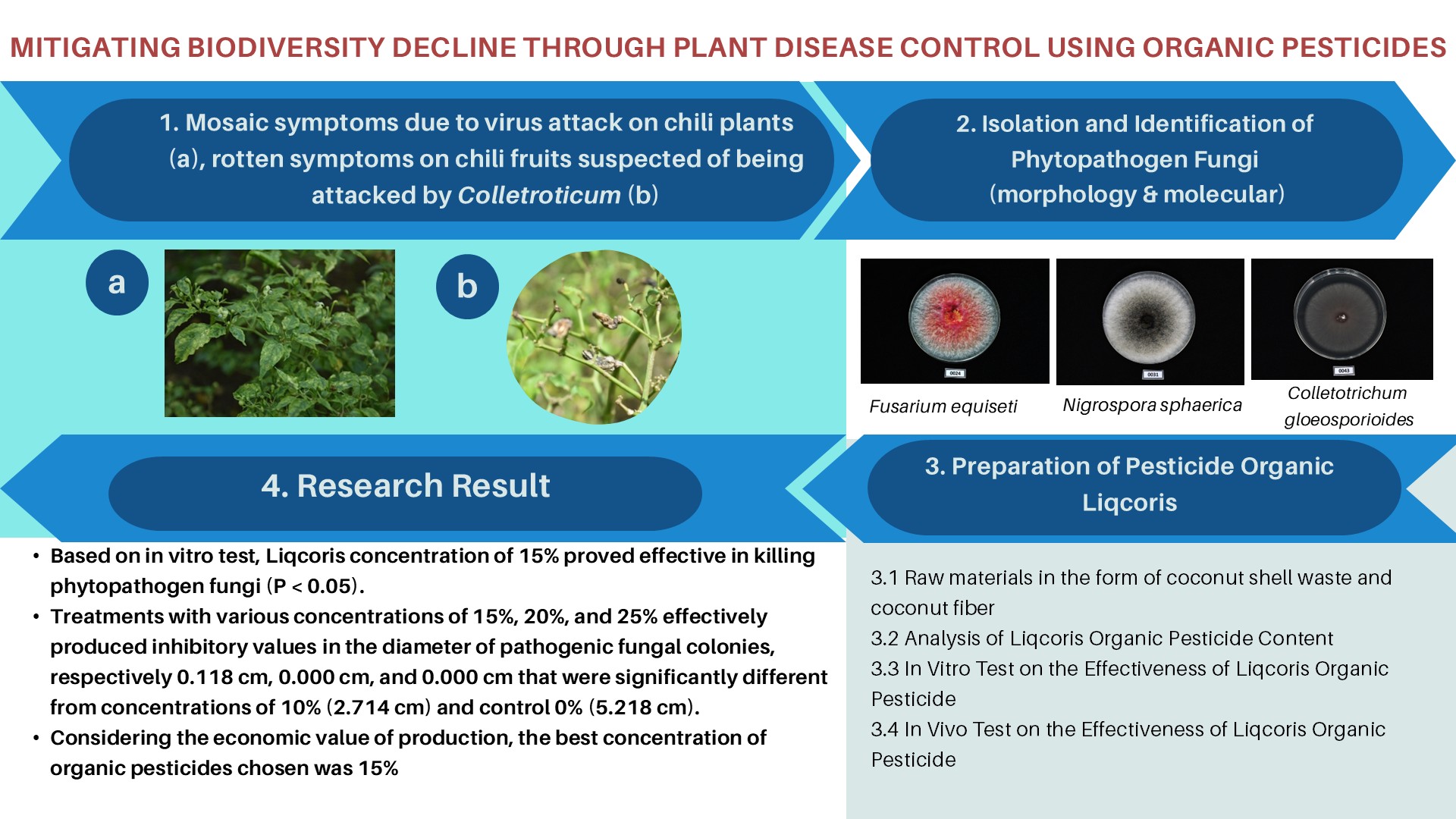
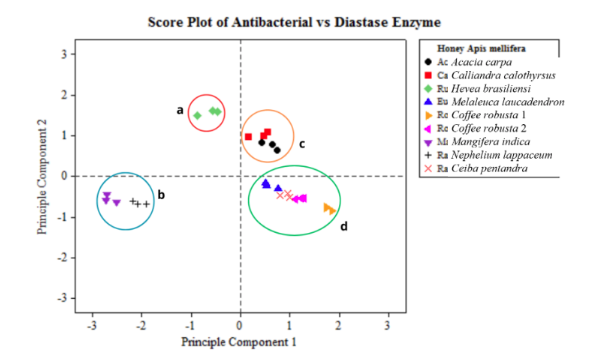
MOLECULAR IDENTIFICATION AND PHYLOGENETIC RELATIONSHIP AMONG LOCAL, SANGKURIANG, AND AFRICAN CATFISH BASED ON RAPD MARKER
Downloads
Catfish species are commercially cultivated freshwater fish by fish farmer. Formerly, only one species of catfish had been cultivated, namely local catfish (walking catfish) Clarias batrachus. However, since 1985, local catfish was replaced by a new commodity from Africa called African catfish (C. gariepinus). In 2004, BBPBAT Sukabumi had introduced a new strain of catfish, namely Sangkuriang catfish. Taxonomic analysis needed, especially for Sangkuriang catfish using molecular marker, in order to get a clearer picture about its genetics departure from their parental, so their taxonomic status and relationships with local and African catfish is clearer.  One of the molecular markers is randomly amplified polymorphic DNA (RAPD) markers. Species status defined based on the presence and absence of specific band on each catfish. Phylogenetics relationships was inferred from phylogenetic tree which was built using UPGMA tool as implemented in NTSYS software based on the similarity of RAPD band pattern. The result showed that Sangkuriang and local catfish had specific RAPD markers, while African catfish was not. This means that all RAPD markers of African catfish were shared with those of two others catfish. Local catfish phylogenetically distantly related to African and Sakuriang catfish. Only one individual of African catfish (D4) genetically close related to Sangkuriang catfish while the rest individuals of African catfish genetically closed related to green catfish. This means that African catfish showed very divergence genetic constituent.Â
Â
Key words: catfish, molecular identification, phylogenetic relationships, RAPDDownloads
Authors who publish with this journal agree with the following terms:
- Authors retain copyright and grant the journal right of first publication, with the work 1 year after publication simultaneously licensed under a Creative Commons attribution-noncommerical-noderivates 4.0 International License that allows others to share, copy and redistribute the work in any medium or format, but only where the use is for non-commercial purposes and an acknowledgement of the work's authorship and initial publication in this journal is mentioned.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See The Effect of Open Access).