INTRA-SPESIFIC DIVERSITY OF BUTTERFLY PEA (Clitoria ternatea L.) REVEALED BY ISSR WITH INVARIABLE ITS RECORDS
Article Highlights:
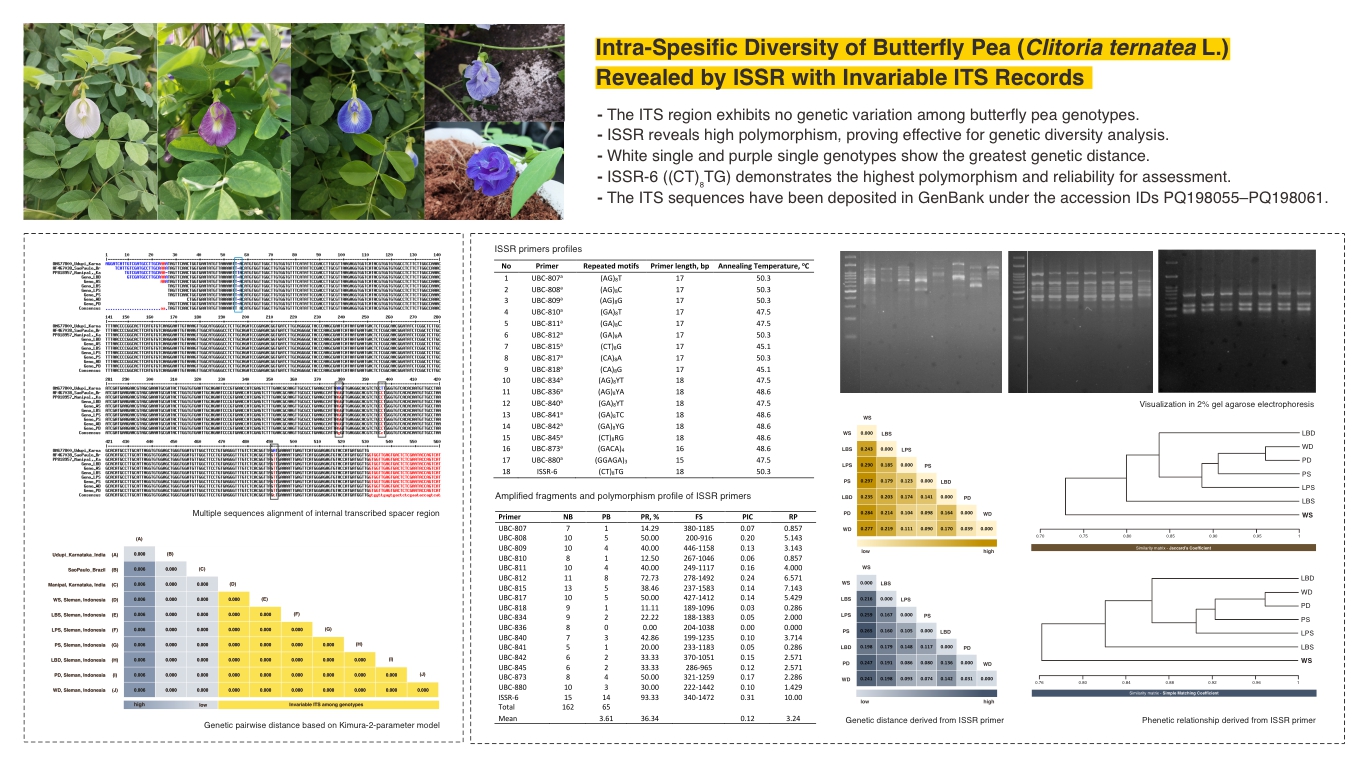
- The ITS region exhibits no genetic variation among butterfly pea genotypes.
- ISSR reveals high polymorphism, proving effective for genetic diversity analysis.
- White single and purple single genotypes show the greatest genetic distance.
- ISSR-6 ((CT)₈TG) demonstrates the highest polymorphism and reliability for assessment.
- The ITS sequences have been deposited in GenBank under the accession IDs PQ198055–PQ198061.
ABSTRACT
Clitoria ternatea L., a perennial plant in the Fabaceae, is recognized for its resilience in tropical climates and its diverse applications in both culinary and medicinal fields. However, the limited exploration of its genetic diversity constrains breeding efforts aimed at improving desirable traits. This limitation highlights the need to optimize selection strategies, identify superior genotypes, and preserve valuable genetic resources for long-term conservation and crop enhancement. This study aimed to explore genetic variation using molecular markers to analyze C. ternatea genotypes based on petal architecture and color differences. To assess the genetic diversity of C. ternatea, Sanger sequencing of the Internal Transcribed Spacer (ITS) region and Inter-Simple Sequence Repeat (ISSR) markers were applied to seven wild populations from Sleman, Yogyakarta. The ITS region exhibited no genetic variation, indicating its conserved nature and limited ability to differentiate genotypes. In contrast, ISSR markers effectively detected genetic variation, identifying 62 polymorphic fragments out of 162 total bands. The highest genetic distance (0.297) was observed between the WS and PS genotypes, whereas the double-petal genotypes (WD and PD) displayed the closest phenetic relationship. Among the ISSR primers, UBC-808, UBC-812, and ISSR-6 exhibited high PIC and RP values, confirming their reliability in genetic diversity analysis. These results underscore the utility of ISSR markers as a robust tool for genetic diversity assessment, offering valuable insights for breeding programs and germplasm conservation in C. ternatea.
Downloads
INTRODUCTION
Clitoria ternatea L. is a perennial plant from the Fabaceae, thrives in tropical climates and demonstrates resilience to environmental disturbances. It is widely recognized for its traditional Ayurvedic medicine and culinary applications(Ashraf et al., 2023);(Maneechot et al., 2023). The flowers are used as supplements to enhance cognitive function(Oguis et al., 2019)and serve as a natural food coloring in rice cakes, tea, and various desserts(Chusak et al., 2018).
Additionally, flower extracts have been reported to exhibit antibacterial, antioxidant, and antidiabetic properties(Rajamanickam et al., 2015);(Jeyaraj et al., 2022).
Morphologically, C. ternatea flowers are solitary, grow in the axils, and have a characteristic papilionaceous structure. This species exhibits notable variations in petal morphology, with flowers having either single or double layers of petals. In the single-layer type, the flower follows a papilionaceous structure, consisting of a standard petal (the largest), two wing petals, and two keel petals. In contrast, the double-layer type is characterized by enlarged wing and keel petals that reach the size of the standard petal, creating a layered appearance. The colors were distinct, ranging from white to blue and purple, corresponding to differences in anthocyanin composition ((Suarna & Wijaya, 2021);(Surya et al., 2022)).
Genetic variation plays a vital role in shaping species’ traits and their capacity to adapt to changes in the environment(Mora et al., 2005). Molecular data-based identification techniques are particularly effective in describing species because they rely on stable genetic markers that are not influenced by environmental factors ((Loureiro & Malfeito-Ferreira, 2006);(Elufioye & Badal, 2017)). A frequently used marker in genetic research is the Internal Transcribed Spacer (ITS) region, which is located between the 16S, 5.8S, and 26S regions of rDNA (Froeschke & Von Der Heyden 2014). Several studies have demonstrated the success of using ITS regions for assessing plant genetic diversity at the inter- and intraspecific levels ((Tripathi et al., 2013);(Sokołowska et al., 2022);(Pere et al., 2023)). ITS1 is generally more variable than ITS2 due to the presence of variable repeat units, making it particularly useful for species discrimination and effective in distinguishing various species in Fabaceae ((Nolan & Cribb, 2005);(Vilas et al., 2005);(, 2010)).
DNA-based molecular markers are other ideal tools for evaluating genetic diversity, breeding- related traits, and other mobile segments due to their neutrality and practicality(Bishoyi et al., 2014)(Hasan et al., 2021)(Aliabadi et al., 2023)(Nidianti et al., 2023)(Rosyidi et al., 2023). Inter Simple Sequence Repeats (ISSR) are a molecular marker technique that utilizes microsatellite sequences as primers to amplify regions between tandem repeats. ISSR markers are widely used due to their reliability, cost-effectiveness, rapid application, and high sensitivity in genetic analysis ((McGlaughlin et al., 2002);(Sarwat, 2012);(Rahali et al., 2022)), and has been widely used to identify intraspecific variation ((Zebarjadi et al., 2016);(Nilkanta et al., 2017);(Verma et al., 2017);(Abdelaziz et al., 2020);(Yusuf & Daryono, 2021);(Devi et al., 2024);(Ahmed et al., 2022-12-23)). ISSR cover a broad portion of the genome, include numerous polymorphic loci, and do not necessitate prior genomic knowledge(Liu et al., 2015);(Tiwari et al., 2016).
Given the considerable variability in petal color and structural traits among C. ternatea genotypes, this study has been implemented to identify genetic variations and expand the knowledge base for breeding programs. Documenting this diversity is crucial for preserving genetic material that may otherwise be lost through breeding and cultivation, as well as for ensuring the continuity of information regarding species richness and existence.
MATERIALS AND METHODS
Genomic DNA Extraction
Seven distinct variants of C. ternatea were collected from a wild population in Sleman, Indonesia, each exhibiting different petal architecture and colors differences. Genomic DNA was extracted from 100 mg of frozen leaf samples with the Genomic DNA Mini Kit Plant (Geneaid, Taiwan). The concentration and purity of the DNA were evaluated using a NanoDrop Lite Plus spectrophotometer (Thermo Fisher Scientific, USA), with acceptable purity from 1.8 to 2.0(Green & Sambrook, 2012).
Amplification Process
PCR amplification was performed using the MyTaq HS RedMix (Bioline, US) protocol with several modifications. The total reaction volume was 20 µL, comprising 10 µL of MyTaq HS RedMix, 2 µL of primer, 2 µL of DNA sample at 50 ng/µL, and 6 µL of nuclease-free water. The amplification process was conducted in a T100 Thermal Cycler (Bio-Rad, US) over 40 cycles. The total reaction volume was 20 µL, consisting of 10 µL of MyTaq HS RedMix, 2 µL of primer, 2 µL of DNA sample (50 ng/µL), and 6 µL of nuclease-free water. Amplification was performed using a T100 Thermal Cycler (Bio-Rad, US) with 40 cycles. Each cycle involved a 45-second denaturation step at 95 °C, a 30-second annealing step at 45 - 50 °C, and a 1-minute elongation step at 72 °C. The protocol began with an initial denaturation at 95 °C for 3 minutes and ended with a final extension at 72 °C for 5 minutes. A universal ITS primer pair was employed, comprising ITS-u1 (5’-GGAAGKARAAGTCGTAACAAGG-3’) and ITS-u4 (5’-RGTTTCTTTTCCTCCGCTTA-3’)(Cheng et al., 2016). Details of the eighteen ISSR primers used are provided inTable 1.
| No | Primer | Repeated motifs | Primer length, bp | Annealing Temperature, |
|---|
Abdelaziz MS, Medraoui L, Alami M, Pakhrou O, Makkaoui M, Ould Mohamed Salem Boukhary A, Filali-Maltouf A. 2020. Inter simple sequence repeat markers to assess genetic diversity of the desert date (Balanites aegyptiaca Del.) for Sahelian ecosystem restoration. Sci Rep 10(1):14948. DOI: 10.1038/s41598-020-71835-9 DOI: https://doi.org/10.1038/s41598-020-71835-9
Ahmed AS, Barznjy LGK, Kasnazany SAS, Mohammed DO. 2022 Dec 23. Genetic diversity study of melon native landraces using ISSR markers in Kurdistan Region of Iraq. J Agric Ecol Res Int 23(6):213-7. DOI: 10.9734/jaeri/2022/v23i6512 DOI: https://doi.org/10.9734/jaeri/2022/v23i6512
Ainouche A, Bayer RJ. 1999. Phylogenetic relationships in Lupinus (Fabaceae: Papilionoideae) based on internal transcribed spacer sequences (ITS) of nuclear ribosomal DNA. Am J Bot 86(4):590-607. DOI: 10.2307/2656820 DOI: https://doi.org/10.2307/2656820
Aliabadi F, Bagheri A, Abbasi S, Saeidi H, Blattner FR. 2023. High genetic diversity in an endemic and vulnerable species: Evidence from Astragalus cyclophyllon (Fabaceae) in Iran. Genet Resour Crop Evol 70(7):1999-2008. DOI: 10.1007/s10722-023-01550-7 DOI: https://doi.org/10.1007/s10722-023-01550-7
Ali Z, Ganie SH, Narula A, Sharma MP, Srivastava PS. 2013. Intra-specific genetic diversity and chemical profiling of different accessions of Clitoria ternatea L. Ind Crop Prod 43:768-73. DOI: 10.1016/j.indcrop.2012.07.070 DOI: https://doi.org/10.1016/j.indcrop.2012.07.070
Aristya GR, Kasiamdari R, Arif MF, Amalia F, Pawestri RD. 2019. Comparison of molecular markers ISSR, SSR and RFLP to study the genetic variation of strawberry cultivars. Am J Agric Biol Sci 14(1):61-8. DOI: 10.3844/ajabssp.2019.61.68 DOI: https://doi.org/10.3844/ajabssp.2019.61.68
Ashraf K, Adlin NF, Basri AN, Ahmad W, Sultan S. 2023. The traditional uses, phytochemistry, and pharmacological effects of Clitoria ternatea: A review. Ind J Pharm Edu Res 58(1):1-14. DOI: 10.5530/ijper.58.1.1 DOI: https://doi.org/10.5530/ijper.58.1.1
Bhadkaria A, Gupta N, Narvekar DT, Bhadkariya R, Saral A, Srivastava N, …, Bhagyawant SS. 2020. ISSR-PCR approach as a means of studying genetic variation in moth bean (Vigna aconitifolia (Jacq.) Maréchal). Biocatal Agric Biotechnol 30:101827. DOI: 10.1016/j.bcab.2020.101827 DOI: https://doi.org/10.1016/j.bcab.2020.101827
Bishoyi AK, Pillai VV, Geetha KA, Maiti S. 2014. Assessment of genetic diversity in Clitoria ternatea populations from different parts of India by RAPD and ISSR markers. Genet Resour Crop Evol 61(8):1597-1609. DOI: 10.1007/s10722-014-0145-y DOI: https://doi.org/10.1007/s10722-014-0145-y
Cheng T, Xu C, Lei L, Li C, Zhang Y, Zhou S. 2016. Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Mol Ecol Resour 16(1):138-49. DOI: 10.1111/1755-0998.12438 DOI: https://doi.org/10.1111/1755-0998.12438
Chusak C, Henry CJ, Chantarasinlapin P, Techasukthavorn V, Adisakwattana S. 2018. Influence of Clitoria ternatea flower extract on the in-vitro enzymatic digestibility of starch and its application in bread. Foods 7(7):102. DOI: 10.3390/foods7070102 DOI: https://doi.org/10.3390/foods7070102
Devi NS, Rajwanshi R, Mohapatra KP. 2024. Evaluating the genetic variability in selected accessions of medicinal legume Mucuna pruriens (L) DC through ISSR markers. Ecol Genet Genom 32:100262. DOI: 10.1016/j.egg.2024.100262 DOI: https://doi.org/10.1016/j.egg.2024.100262
Dos Santos Araújo F, Vasconcelos Pacheco M, De Almeida Vieira F, Dos Santos Ferrari C, Cardoso Félix F, Teixeira Das Chagas KP. 2016. ISSR molecular markers for the study of the genetic diversity of Mimosa caesalpiniaefolia Benth. Idesia 34(3):47-52. DOI: 10.4067/S0718-34292016000300007 DOI: https://doi.org/10.4067/S0718-34292016000300007
Elufioye TO, Badal S. 2017. Background to pharmacognosy. In: Badal S, Delgoda R (Editors). Pharmacognosy: Fundamentals, Applications, and Strategies. Amsterdam (NL): Elsevier. p. 3-13. DOI: 10.1016/B978-0-12-802104-0.00001-9 DOI: https://doi.org/10.1016/B978-0-12-802104-0.00001-9
Froeschke G, Von Der Heyden S. 2014. A review of molecular approaches for investigating patterns of coevolution in marine host–parasite relationships. In: Rollinson D (Editor). Advances in Parasitology. 1st Edition. Vol. 84. Chapter 4. Oxford (UK): Elsevier. p. 209-52. DOI: 10.1016/B978-0-12-800099-1.00004-1 DOI: https://doi.org/10.1016/B978-0-12-800099-1.00004-1
Gao T, Yao H, Song J, Liu C, Zhu Y, Ma X, …, Chen S. 2010. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J Ethnopharmacol 130(1):116-21. DOI: 10.1016/j.jep.2010.04.026 DOI: https://doi.org/10.1016/j.jep.2010.04.026
Gebhardt C. 2007. Molecular markers, maps and population genetics. In: Vreugdenhil D, Bradshaw J, Gebhardt C, Govers F, Mackerron DKL, Taylor MA, Ross H (Editors). Potato Biology and Biotechnology: Advances and Perspectives. Oxford (UK): Elsevier. p. 77–89. DOI: 10.1016/B978-044451018-1/50047-6 DOI: https://doi.org/10.1016/B978-044451018-1/50047-6
Green MR, Sambrook J. 2012. Molecular cloning: A laboratory manual. Vol. 1. 4th Edition. New York (US): Cold Spring Harbor Laboratory Press.
Haque I, Bandopadhyay R, Mukhopadhyay K. 2009. Intraspecific variation in Commiphora wightii populations based on Internal Transcribed Spacer (ITS1-5.8S-ITS2) sequences of rDNA. Diversity 1(2):89-101. DOI: 10.3390/d1020089 DOI: https://doi.org/10.3390/d1020089
Hasan N, Choudhary S, Naaz N, Sharma N, Laskar RA. 2021. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J Genet Eng Biotechnol 19(1): 128. DOI: 10.1186/s43141-021-00231-1 DOI: https://doi.org/10.1186/s43141-021-00231-1
Helal H, Sayed AEH, Helal A, El Shaer H. 2023. Genetical studies on some genotypes in chickpea (Cicer arietinum L.) by Gamma Rays and ISSR Marker. Al-Azhar J Agric Res 48(3):475-92. DOI: 10.21608/ajar.2024.227841.1215 DOI: https://doi.org/10.21608/ajar.2024.227841.1215
Jeyaraj EJ, Lim YY, Choo WS. 2022. Antioxidant, cytotoxic, and antibacterial activities of Clitoria ternatea flower extracts and anthocyanin-rich fraction. Sci Rep 12(1):14890. DOI: 10.1038/s41598-022-19146-z DOI: https://doi.org/10.1038/s41598-022-19146-z
Lee SY, Mohamed R, Faridah-Hanum I, Lamasudin DU. 2018. Utilization of the internal transcribed spacer (ITS) DNA sequence to trace the geographical sources of Aquilaria malaccensis Lam. populations. Plant Genet Resour 16(2):103-11. DOI: 10.1017/S1479262117000016 DOI: https://doi.org/10.1017/S1479262117000016
Liu C, Xue GP, Cheng B, Wang X, He J, Liu GH, Yang WJ. 2015. Genetic diversity analysis of Capparis spinosa L. populations by using ISSR markers. Genet Mol Res 14(4):16476-83. DOI: 10.4238/2015.December.9.19 DOI: https://doi.org/10.4238/2015.December.9.19
Loureiro V, Malfeito-Ferreira M. 2006. Dekkera/Brettanomyces spp. In: de W. Blackburn C (Editor). Food spoilage microorganisms. Oxford (UK): Elsevier. p. 354-98. DOI: 10.1533/9781845691417.3.354 DOI: https://doi.org/10.1533/9781845691417.3.354
Maneechot O, Hahor W, Thongprajukaew K, Nuntapong N, Bubaka S. 2023. A natural blue colorant from butterfly pea (Clitoria ternatea) petals for traditional rice cooking. J Food Sci Technol 60(8):2255-64. DOI: 10.1007/s13197-023-05752-w DOI: https://doi.org/10.1007/s13197-023-05752-w
McGlaughlin M, Karoly K, Kaye T. 2002. Genetic variation and its relationship to population size in reintroduced populations of pink sand verbena, Abronia umbellata subsp. breviflora (Nyctaginaceae). Conserv Genet 3(4):411-20. DOI: 10.1023/A:1020507416654 DOI: https://doi.org/10.1023/A:1020507416654
Mora F, Palma-Rojas C, Jara-Seguel P. 2005. Comparación del cariotipo de Eucalyptus globulus y Eucalyptus cladocalyx (Myrtaceae) [Comparison of karyotype of Eucalyptus globulus and Eucalyptus cladocalyx (Myrtaceae)]. Agric Téc 65(1):20-25. DOI: 10.4067/S0365-28072005000100002 DOI: https://doi.org/10.4067/S0365-28072005000100002
Naik A, Mishra SK, Nag A, Soren GK, Panda AK, Panda SK, Panigrahi J. 2020. Cross-genera amplification of Cajanus spp. specific SSR markers in Clitoria ternatea (L.) and their application in genetic diversity studies. Physiol Mol Biol Plants 26(12):2371-90. DOI: 10.1007/s12298-020-00907-x DOI: https://doi.org/10.1007/s12298-020-00907-x
Nidianti E, Subiastuti AS, Yusuf AF, Wibowo WA, Ramadhani PH, Daryono BS. 2023. Biological response and molecular identification of the Begomovirus infection in the new Indonesian melon genotype Kinaya. Pak J Phytopathol 35(2):223-34. DOI: 10.33866/phytopathol.035.02.0939 DOI: https://doi.org/10.33866/phytopathol.035.02.0939
Nilkanta H, Amom T, Tikendra L, Rahaman H, Nongdam P. 2017. ISSR marker based population genetic study of Melocanna baccifera (Roxb.) Kurz: A commercially important bamboo of Manipur, North-East India. Scientifica 2017:1-9. DOI: 10.1155/2017/3757238 DOI: https://doi.org/10.1155/2017/3757238
Nolan MJ, Cribb TH. 2005. The use and implications of ribosomal DNA sequencing for the discrimination of Digenean species. In: Baker JR, Muller R, Rollinson D. Advances in Parasitology. Vol.60. Oxford (UK): Elsevier. p.101-63. DOI: 10.1016/S0065-308X(05)60002-4 DOI: https://doi.org/10.1016/S0065-308X(05)60002-4
Nurhasanah, Hindersah R, Suganda T, Concibido V, Sundari, Karuniawan A. 2023. The first report on the application of ISSR markers in genetic variance detection among butterfly pea (Clitoria ternatea L.) accession in North Maluku Province, Indonesia. Horticulturae 9(9): 1059. DOI: 10.3390/horticulturae9091059 DOI: https://doi.org/10.3390/horticulturae9091059
Oguis GK, Gilding EK, Jackson MA, Craik DJ. 2019. Butterfly pea (Clitoria ternatea), a cyclotide-bearing plant with applications in agriculture and medicine. Front Plant Sci 10:645. DOI: 10.3389/fpls.2019.00645 DOI: https://doi.org/10.3389/fpls.2019.00645
Ojuederie OB, Nkang NA, Odesola KA, Igwe DO. 2020. Genetic diversity assessment of winged bean (Psophorcarpus tetragonolobus) accessions revealed by Inter-Simple Sequence Repeat (ISSR) markers. J Plant Biol Crop Res 3(1):1014.
Pere K, Mburu K, Muge EK, Wagacha JM, Nyaboga EN. 2023. Molecular discrimination and phylogenetic relationships of Physalis species based on ITS2 and rbcL DNA barcode sequence. Crops 3(4):302-19. DOI: 10.3390/crops3040027 DOI: https://doi.org/10.3390/crops3040027
Poczai P, Hyvönen J. 2010. Nuclear ribosomal spacer regions in plant phylogenetics: Problems and prospects. Mol Biol Rep 37(4):1897-1912. DOI: 10.1007/s11033-009-9630-3 DOI: https://doi.org/10.1007/s11033-009-9630-3
Porth I, El-Kassaby Y. 2014. Assessment of the genetic diversity in forest tree populations using molecular markers. Diversity 6(2): 283–295. DOI: 10.3390/d6020283 DOI: https://doi.org/10.3390/d6020283
Qahtan AA. 2021. Genetic diversity and structure analysis of a worldwide collection of Faba bean (Vicia faba) genotypes using ISSR markers. IJAB 25(03):683-91. DOI: 10.17957/IJAB/15.1717 DOI: https://doi.org/10.17957/IJAB/15.1717
Rahali N, Yangui I, Boussaid M, Messaoud C. 2022. Assessment of genetic diversity and population structure of the endemic Hertia cheirifolia (L.) Kuntze based on ISSR and SRAP molecular markers. Biologia 77(12):3429-39. DOI: 10.1007/s11756-022-01166-9 DOI: https://doi.org/10.1007/s11756-022-01166-9
Rajamanickam M, Kalaivanan P, Sivagnanam I. 2015. Evaluation of anti-oxidant and anti-diabetic activity of flower extract of Clitoria ternatea L. J App Pharm Sci 5(8):131-8. DOI: 10.7324/JAPS.2015.50820 DOI: https://doi.org/10.7324/JAPS.2015.50820
Reddy P, Sabara P, Padhiyar SM, Kulkarni GU, Kheni JV, Tomar RS. 2022. Genetic diversity of groundnut (Arachis hypogaea L.) revealed by RAPD and ISSR markers. Annals of Arid Zone 60(3):109-15.
Rosyidi IN, Yusuf AF, Sahroni M, Daryono BS. 2023. Detection of transposon gene in lurik peanuts (Arachis hypogaea var. lurik L.) with AhMITEs analysis. Int J Adv Sci Eng Inf Technol 13(4):1349-54. DOI: 10.18517/ijaseit.13.4.18269 DOI: https://doi.org/10.18517/ijaseit.13.4.18269
Sarwat M. 2012. ISSR: A reliable and cost-effective technique for detection of DNA polymorphism. In: Sucher NJ, Hennell JR, Carles MC (Editors). Plant DNA Fingerprinting and Barcoding. Totowa (US): Humana Press. p.103-21. DOI: 10.1007/978-1-61779-609-8_9 DOI: https://doi.org/10.1007/978-1-61779-609-8_9
Shiran B, Amirbakhtiar N, Kiani S, Mohammadi Sh, Sayed-Tabatabaei BE, Moradi H. 2007. Molecular characterization and genetic relationship among almond cultivars assessed by RAPD and SSR markers. Sci Hortic 111(3):280-92. DOI: 10.1016/j.scienta.2006.10.024 DOI: https://doi.org/10.1016/j.scienta.2006.10.024
Sokołowska J, Fuchs H, Celiński K. 2022. Assessment of ITS2 region relevance for taxa discrimination and phylogenetic inference among Pinaceae. Plants 11(8):1078. DOI: 10.3390/plants11081078 DOI: https://doi.org/10.3390/plants11081078
Suarna IW, Wijaya IMS. 2021. Butterfly pea (Clitoria ternatea L.: Fabaceae) and its morphological variations in Bali. J Trop Biodiv Biotechnol 6(2):63013. DOI: 10.22146/jtbb.63013 DOI: https://doi.org/10.22146/jtbb.63013
Sultana S, Lim YP, Bang J-W, Choi H-W. 2011. Internal transcribed spacer (ITS) and genetic variations in Lilium native to Korea. Hortic Environ Biotechnol 52(5):502-10. DOI: 10.1007/s13580-011-0050-7 DOI: https://doi.org/10.1007/s13580-011-0050-7
Surya D, Rajamani K, Uma D. 2022. Morphological characterization and assessment of anthocyanin in three different genotypes of Clitoria ternatea L. Pharma Innovation Journal 11(7):2388-92.
Tiwari G, Singh R, Singh N, Choudhury DR, Paliwal R, Kumar A, Gupta V. 2016. Study of arbitrarily amplified (RAPD and ISSR) and gene targeted (SCoT and CBDP) markers for genetic diversity and population structure in Kalmegh [Andrographis paniculata (Burm. f.) Nees]. Ind Crops Prod 86:1-11. DOI: 10.1016/j.indcrop.2016.03.031 DOI: https://doi.org/10.1016/j.indcrop.2016.03.031
Tripathi AM, Tyagi A, Kumar A, Singh A, Singh S, Chaudhary LB, Roy S. 2013. The Internal Transcribed Spacer (ITS) region and trnhH-psbA are suitable candidate loci for DNA barcoding of tropical tree species of India. PLoS ONE 8(2):e57934. DOI: 10.1371/journal.pone.0057934 DOI: https://doi.org/10.1371/journal.pone.0057934
Vander Stappen J, De Laet J, Gama-López S, Van Campenhout S, Volckaert G. 2002. Phylogenetic analysis of Stylosanthes (Fabaceae) based on the internal transcribed spacer region (ITS) of nuclear ribosomal DNA. Plant Syst Evol 234(1):27-51. DOI: 10.1007/s00606-002-0193-1 DOI: https://doi.org/10.1007/s00606-002-0193-1
Verma KS, Ul Haq S, Kachhwaha S, Kothari SL. 2017. RAPD and ISSR marker assessment of genetic diversity in Citrullus colocynthis (L.) Schrad: A unique source of germplasm highly adapted to drought and high-temperature stress. 3 Biotech 7(5):288. DOI: 10.1007/s13205-017-0918-z DOI: https://doi.org/10.1007/s13205-017-0918-z
Vijayan K, Chatterjee SN. 2003. ISSR profiling of Indian cultivars of mulberry (Morus spp.) and its relevance to breeding programs. Euphytica 131(1):53-63. DOI: 10.1023/A:1023098908110 DOI: https://doi.org/10.1023/A:1023098908110
Vilas R, Criscione CD, Blouin MS. 2005. A comparison between mitochondrial DNA and the ribosomal internal transcribed regions in prospecting for cryptic species of platyhelminth parasites. Parasitology 131(06):839. DOI: 10.1017/S0031182005008437 DOI: https://doi.org/10.1017/S0031182005008437
Wu C-T, Hsieh C-C, Lin W-C, Tang C-Y, Yang C-H, Huang Y-C, Ko Y-J. 2013. Internal transcribed spacer sequence-based identification and phylogenic relationship of I-Tiao-Gung originating from Flemingia and Glycine (Leguminosae) in Taiwan. J Food Drug Anal 21(4):356-62. DOI: 10.1016/j.jfda.2013.08.002 DOI: https://doi.org/10.1016/j.jfda.2013.08.002
Xu B, Zeng X-M, Gao X-F, Jin D-P, Zhang L-B. 2017. ITS non-concerted evolution and rampant hybridization in the legume genus Lespedeza (Fabaceae). Sci Rep 7(1):40057. DOI: 10.1038/srep40057 DOI: https://doi.org/10.1038/srep40057
Yeotkar SD, Malode SN, Waghmare VN, Thakre P. 2011. Genetic relationship and diversity analysis of Clitoria ternatea variants and Clitoria biflora using random amplified polymorphic DNA (RAPD) markers. Afr J Biotechnol 10(79):18065-70. DOI: 10.5897/AJB11.1423 DOI: https://doi.org/10.5897/AJB11.1423
Yusuf AF, Alfiyani A, Salsabila TAS, Kusnanda PS, Hidayati IN, Daryono BS. 2023. Assessment of maturity stage and stability of new Indonesian melon cultivar ‘Melona’ based on ISSR markers and morphological characteristics. Biodiversitas 24(1). DOI: 10.13057/biodiv/d240137 DOI: https://doi.org/10.13057/biodiv/d240137
Yusuf AF, Daryono BS. 2021. Studies of genetic and morphological characteristics of Indonesian melon (Cucumis melo L. ‘Hikapel’) germplasm. Int J Adv Sci Eng Inf Technol 11(5):2023-30. DOI: 10.18517/IJASEIT.11.5.14047 DOI: https://doi.org/10.18517/ijaseit.11.5.14047
Zebarjadi A, Ahmadvandi HR, Kahrizi D, Cheghamirza K. 2016. Assessment of genetic diversity by application of Inter Simple Sequence Repeat (ISSR) primers on Iranian Harmal (Peganum harmala L.) germplasm as an important medicinal plant. J Appl Biotechnol Rep 3(3):441-45.
Zhao L, Feng S, Tian J, Wei A, Yang T. 2018. Internal transcribed spacer 2 ( ITS 2) barcodes: A useful tool for identifying Chinese Zanthoxylum. Appl Plant Sci 6(6):e01157. DOI: 10.1002/aps3.1157 DOI: https://doi.org/10.1002/aps3.1157
Copyright (c) 2025 Adib Fakhruddin Yusuf, Vida Rahma Latifah, Vivi Indah Nurcahyati, Anggun Diyan Nurhasanah, Adristi Shafa Widyasari, Ananto Puradi Nainggolan, Aldy Riau Wansyah Hasibuan, Madyan Akmal Hidayat, Karmilah, Arini Dian Pratiwi, Rindu Aurantika, Muslifah Hasanah, Ganies Riza Aristya, Niken Satuti Nur Handayani, Tuty Arisuryanti, Indra Lesmana, Budi Setiadi Daryono

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Authors who publish with this journal agree with the following terms:
- Authors retain copyright and grant the journal right of first publication, with the work 1 year after publication simultaneously licensed under a Creative Commons attribution-noncommerical-noderivates 4.0 International License that allows others to share, copy and redistribute the work in any medium or format, but only where the use is for non-commercial purposes and an acknowledgement of the work's authorship and initial publication in this journal is mentioned.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See The Effect of Open Access).