SURVEILLANCE OF β-LACTAMASE GENES OF SALMONELLA FROM CHICKEN IN WET MARKETS OF METRO MANILA, PHILIPPINES
Article Highlights
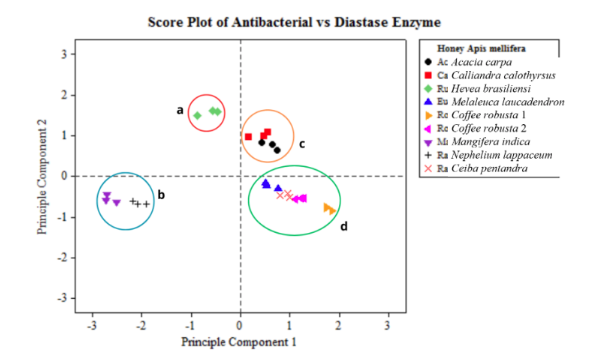
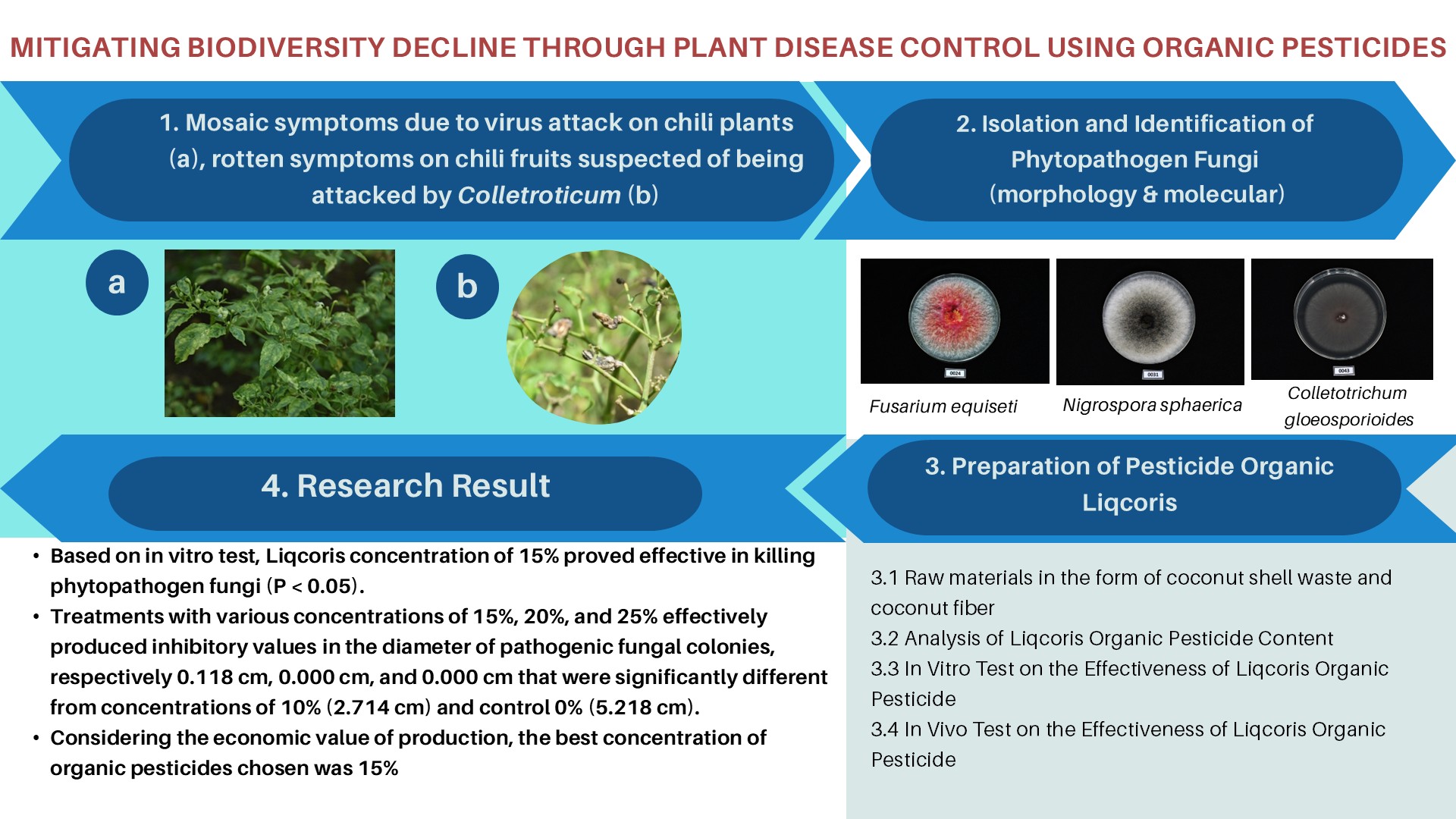
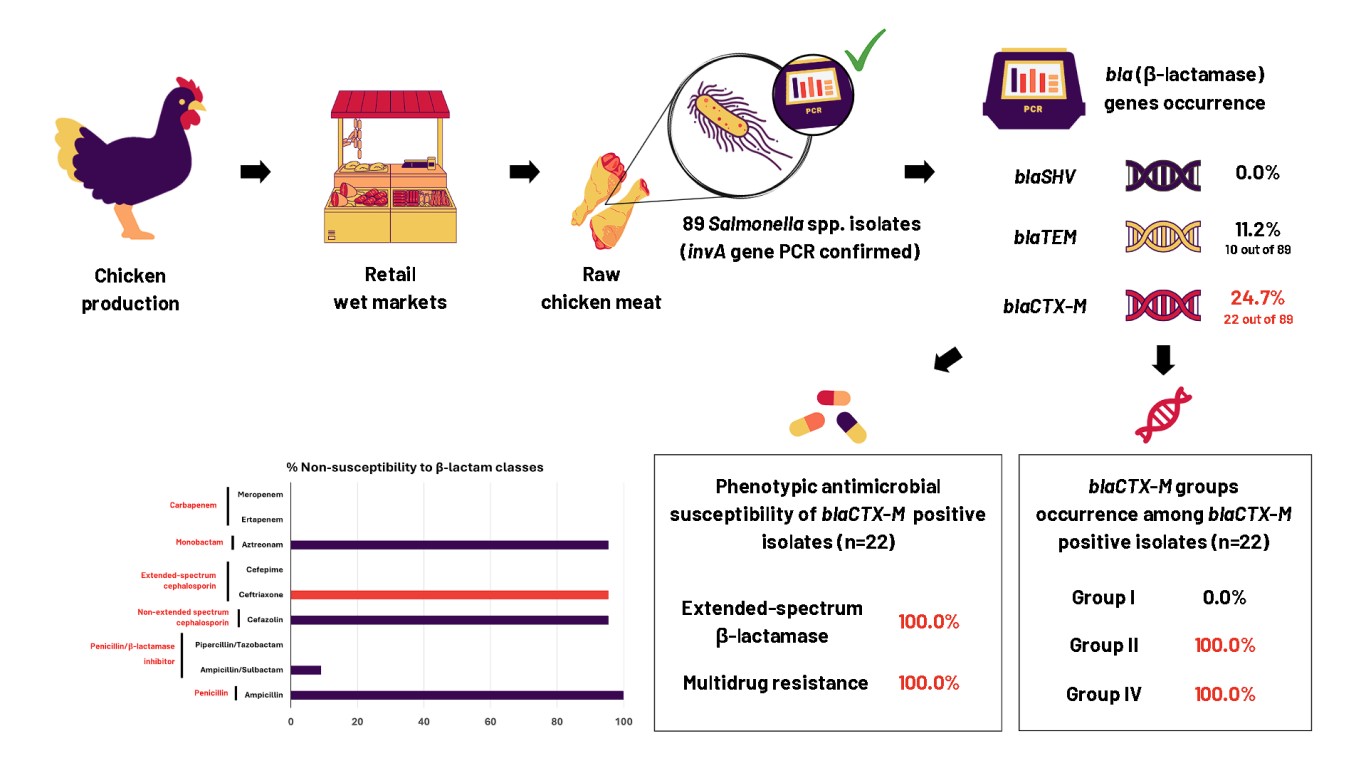
- blaCTX-M gene in 24.7% while blaTEM gene in 11.2% of Salmonella isolates.
- Coexistence of blaCTX-M groups II and IV in all blaCTX-M-positive isolates.
- Dominance of blaCTX-M corroborated with phenotypic β-lactam resistances.
- blaCTX-M-positive isolates had extended-spectrum β-lactamase and multidrug resistance.
Abstract
Salmonella sp. is a foodborne pathogenic bacterium causing millions of cases with hundred thousand death incidents. Infection by Salmonella can diversely manifest as gastroenteritis, bacteremia, and enteric fever. Salmonella can be transmitted through direct consumption of contaminated foods especially animal-based foods, such as chicken meat and its derivatives. Over the years, antimicrobial resistance (AMR) and diverse β-lactamase (bla) gene-carrying Salmonella strains have been reported. These facts are alarming given that cephalosporins are a major class of β-lactam antibiotics used in clinical settings. Hence, the main objective of this study was to molecularly detect the occurrence of different bla genes by Polymerase Chain Reaction (PCR) and profile the phenotypic antimicrobial susceptibility of Salmonella collected from various chicken sample types in wet markets of Metro Manila, Philippines. Of the 89 Salmonella isolates, blaCTX-M had the highest occurrence, detected in 22 isolates (24.7%), while blaTEM was detected in 10 isolates (11.2%). Genotypic and phenotypic resistance corroboration was observed in nearly all blaCTX-M-positive Salmonella tested, with all strains showing resistance to ampicillin and nitrofurantoin (100%) and 21 out of 22 (95.5%) exhibiting resistance to both non-extended and extended-spectrum cephalosporins. In addition, blaCTX-M groups II and IV genes were co-detected and multidrug resistance (MDR) profiles were also observed in all blaCTX-M-positive isolates. The high AMR patterns of Salmonella isolates suggest potential threats to food safety and public health. Additionally, the corroboration of phenotypic and genotypic resistance and the high occurrence of MDR among Salmonella isolates highlight the importance of continued surveillance of AMR genes and regulation of antimicrobial use to combat AMR.
Downloads
INTRODUCTION
Salmonella sp. is a Gram-negative, facultative anaerobic, mesophilic, rod-shaped foodborne bacterium that belongs to the family Enterobacteriaceae(Ethelberg et al., 2014). Given that poultry is a significant reservoir for Salmonella, the risk of transmission increases through improper handling, trade, or slaughter of raw chicken(Eng et al., 2015). Infection can occur through the fecal-oral route, involving the ingestion of contaminated food, especially animal-based foods, such as chicken meat, or through water containing Salmonella(Eng et al., 2015). Due to their easy transmission from raw/improperly cooked meat, the high consumption of chicken in the Philippines can pose a risk for Salmonella infection. In the Philippines, the average annual poultry consumption per capita in the period of 2009-2018 was 13.4 kg(D.A.-B.A.I., 2022).
Although Salmonella consists of only two species, enterica and bongori, with S. enterica containing six subspecies, most infections are caused by S. enterica subsp. enterica(Desai et al., 2013). Infections can manifest as enteric fever, gastroenteritis, bacteremia, or a chronic carrier state depending on the Salmonella serovars. Enteric fever is caused by S. Typhi or S. Paratyphi A, B, and C which are typhoidal Salmonella. All other Salmonella strains are designated as nontyphoidal Salmonella (NTS), which causes mild infections, including gastroenteritis. Although typhoidal Salmonella is more likely to follow a human-to-human transmission route, NTS transmission is more likely associated with animal reservoirs, such as contaminated chicken meat. The most common serovars responsible for NTS are S. Enteritidis, S. Typhimurium, and S. Newport. The symptoms of enteric fever include headache, abdominal pain, diarrhea/constipation, fever, rose spots, and in severe cases, bloody diarrhea. Conversely, the symptoms of gastroenteritis are usually self-limiting and include headache, abdominal cramps, vomiting, non-bloody diarrhea, nausea, and muscle ache(Eng et al., 2015).
NTS and typhoidal Salmonella may eventually lead to bacteremia and a chronic carrier state. Bacteremia occurs when Salmonella penetrates the intestinal barrier and invades the bloodstream, while a chronic carrier state is characterized by fecal shedding of Salmonella more than one year after the acute stage of Salmonella infection(Eng et al., 2015). In 2017, there were approximately 14.3 million cases of typhoidal Salmonella and approximately 95.6 million cases of NTS globally(Stanaway et al., 2019). Approximately 136,000 died from typhoidal Salmonella in the same year, and 128,000 died from NTS(Stanaway et al., 2019). In the Philippines,(Santos et al., 2020)found S. enterica in meat sold at wet markets in Metro Manila, highlighting the need to investigate the presence of bla genes among S. enterica isolates.
Due to numerous Salmonella-related deaths annually, antibiotic treatment becomes important for treatments against invasive diseases. Extended-spectrum cephalosporins (ESC) are some of the antibiotics used to treat Salmonella infections(Calayag et al., 2021). The treatments for NTS also include ciprofloxacin and ceftriaxone, and for severe complications, may include cefixime and cefotaxime(Gut et al., 2018). Cephalosporins rely on cell wall synthesis interference through the inhibition of transpeptidases, which causes bacterial cell lysis and death(Cantón, 2007). However, antimicrobial resistance (AMR) threatens the efficacy of these treatments. Some Salmonella strains have acquired resistance to these antibiotics because of their ability to produce β-lactamase enzymes that hydrolyze class A β-lactam antibiotics, including cephalosporins(Cantón et al., 2012).
β-lactamase enzymes, such as temoneira (TEM), cefotaximase (CTX-M), and sulfhydryl variable (SHV) are encoded by blaTEM, blaCTX-M,and blaSHVgenes, respectively, and are among the most common ESBL genes(Ejaz et al., 2021). Additionally, blaCTX-Mgenes are highly diverse and prevalent among Enterobacteriaceae, due to transmissions, mutations, and recombinations. They include numerous groups that confer resistance to different generations of cephalosporins, primarily against cefotaxime and ceftriaxone, with some enhanced variants capable of acting even against ceftazidime(, 2008). Some extended-spectrum β-lactamases (ESBLs) also have the capability to hydrolyze broad-spectrum third- and fourth-generation cephalosporins(Cantón, 2007), posing a significant threat in clinical treatment settings. Determining the occurrence of these genes and phenotypic resistance of S. enterica is important for understanding its transmission and dynamics. Hence, this study determined the occurrence of blaTEM, blaCTX-M, and blaSHV, in S. enterica isolated from raw chicken from wet markets in Metro Manila, Philippines. Moreover, this study is among the first in the Philippines to detect the simultaneous occurrence of blaCTX-Mgene groups and determined the phenotypic antimicrobial susceptibility profile of S. enterica possessing those gene groups.
MATERIALS AND METHODS
Salmonella Isolates
Following standard procedures under ISO 6579-1:2017(Standardization, 2017)and Santos et al. (2020), 25 g of raw chicken meat samples from leg, thigh, and breast parts were aseptically minced and placed in sterile Whirl-Pak® bags. Then, 225 mL of buffered peptone water (BPW) (BD Diagnostics System, NJ, USA) was added, followed by homogenization for 30 seconds and incubation at 37 °C for 18-24 hours. Subsequently, 100 μL of the BPW culture was transferred to 9 mL of Rappaport Vassiliadis (RV) (BD Diagnostics System, NJ, USA) and incubated at 42 °C for 18-24 hours. The resulting RV cultures were then streaked on xylose lysine deoxycholate (XLD) agar (BD Diagnostics System, NJ, USA) and incubated at 37 °C for 18-24 hours. Black colonies on red XLD agar were then subcultured on nutrient agar (NA) (BD Diagnostics System, NJ, USA) plates and incubated at 37 °C for 18-24 hours. The resulting colonies were then subjected to DNA extraction, Salmonella confirmation,
Barroga TRM, Morales RG, Benigno CC, Castro SJM, Caniban MM, Cabullo MFB, Dorado-Garcia A. 2020. Antimicrobials used in backyard and commercial poultry and swine farms in the Philippines: A qualitative pilot study. Front Vet Sci 7:329. DOI: https://doi.org/10.3389/fvets.2020.00329
Calayag AMB, Paclibare PAP, Santos PDM, Bautista CAC, Rivera WL. 2017. Molecular characterization and antimicrobial resistance of Salmonella enterica from swine slaughtered in two different types of Philippine abattoir. Food Microbiol 65:51-6. DOI: https://doi.org/10.1016/j.fm.2017.01.016
Calayag AMB, Widmer KW, Rivera WL. 2021. Antimicrobial susceptibility and frequency of bla and qnr genes in Salmonella enterica isolated from slaughtered pigs. Antibiotics 10(12):1442. DOI: https://doi.org/10.3390/antibiotics10121442
Cantón R. 2007. Chapter 22: Epidemiology and evolution of beta-lactamases. In: Baquero F, Nombela C, Cassell G, Gutiérrez-Fuentes J (Editors). Evolutionary Biology of Bacterial and Fungal Pathogens. Washington DC (US): Wiley Online Library. p. 249-70.
Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: Origin and diffusion. Front Microbiol 3:110. DOI: 10.3389/fmicb.2012.00110 DOI: https://doi.org/10.3389/fmicb.2012.00110
Chiu CH, Ou JT. 1996. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol 34(10):2619-22. DOI: https://doi.org/10.1128/jcm.34.10.2619-2622.1996
[CLSI] Clinical and Laboratory Standards Institute. 2022. Performance standards for antimicrobial susceptibility testing, 32nd ed. CLSI Guideline M100.
Coculescu B-l, Palade A-M, Delcaru C, Pircalabioru G, Berteșteanu Ș. 2015. Phenotypic and genotypic characterization of Salmonella Typhimurium strains producing extended-spectrum beta-lactamases (ESBLs) isolated from children (under 4 years of age) with diarrhea in Romania. Rom Biotechnol Lett 20(3):10478-85.
Cruz MC, Hedreyda CT. 2017. Detection of plasmid-borne β-lactamase genes in extended-spectrum β-lactamase (ESBL) and non-ESBL-producing Escherichia coli clinical isolates. Philipp J Sci 146(2):167-75.
[DA-BAI] Department of Agriculture-Bureau of Animal Industry. 2022. The Philippine Poultry Broiler Industry Roadmap (2022-2040). Quezon City (PH).
Deekshit VK, Srikumar S. 2022. “To be, or not to be”-The dilemma of “silent” antimicrobial resistance genes in bacteria. J Appl Microbiol 133(5):2902-14. DOI: https://doi.org/10.1111/jam.15738
Desai PT, Porwollik S, Long F, Cheng P, Wollam A, Bhonagiri-Palsikar V, McClelland M. 2013. Evolutionary Genomics of Salmonella enterica Subspecies. mBio 4(2). DOI: https://doi.org/10.1128/mBio.00198-13
Ejaz H, Younas S, Abosalif KOA, Junaid K, Alzahrani B, Alsrhani A, Hamam SSM. 2021. Molecular analysis of blaSHV, blaTEM, and blaCTX-M in extended-spectrum β-lactamase producing Enterobacteriaceae recovered from fecal specimens of animals. PLoS ONE 16(1):e0245126. DOI: https://doi.org/10.1371/journal.pone.0245126
Eng SK, Pusparajah P, Ab Mutalib NS, Ser HL, Chan KG, Lee LH. 2015. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci 8(3):284-93. DOI: https://doi.org/10.1080/21553769.2015.1051243
Ethelberg S, Mølbak K, Josefsen MH. 2014. Bacteria: Salmonella non-Typhi. In: Motarjemi Y (Editor). Encyclopedia of Food Safety. Waltham (US): Academic Press. p. 501-14. DOI: https://doi.org/10.1016/B978-0-12-378612-8.00112-8
Ferreira J, Penha Filho R, Andrade L, Berchieri A, Darini A. 2014. Detection of chromosomal blaCTX-M-2 in diverse Escherichia coli isolates from healthy broiler chickens. Clin Microbiol Infect 20(10):O623-O626. DOI: https://doi.org/10.1111/1469-0691.12531
Ghazaei C. 2018. Phenotypic and molecular detection of β-lactamase genes blaTEM, blaCTX, and blaSHV produced by Salmonella spp. isolated from poultry meat. Gene Cell Tissue 5(4):e84367. DOI: 10.5812/gct.84367 DOI: https://doi.org/10.5812/gct.84367
Gundran RS, Cardenio PA, Villanueva MA, Sison FB, Benigno CC, Kreausukon K, Punyapornwithaya V. 2019. Prevalence and distribution of blaCTX-M, blaSHV, blaTEM genes in extended-spectrum β-lactamase-producing E. coli isolates from broiler farms in the Philippines. BMC Vet Res 15(2019):1-8. DOI: https://doi.org/10.1186/s12917-019-1975-9
Gut AM, Vasiljevic T, Yeager T, Donkor ON. 2018. Salmonella infection: Prevention and treatment by antibiotics and probiotic yeasts: A review. Microbiology 164(11):1327-44. DOI: https://doi.org/10.1099/mic.0.000709
He D, Partridge SR, Shen J, Zeng Z, Liu L, Rao L, Liu JH. 2013. CTX-M-123, a novel hybrid of the CTX-M-1 and CTX-M-9 group β-lactamases recovered from Escherichia coli isolates in China. Antimicrob Agents Chemother 57(8):4068-71. DOI: https://doi.org/10.1128/AAC.00541-13
Imperial IC, Pabustan PM, Valencia KA, Nicdao MA, Ibana J. 2022. Emergence of resistance genes in fecal samples of antibiotic-treated Philippine broilers emphasizes the need to review local farming practices. Trop Biomed 39(1):150-9. DOI: https://doi.org/10.47665/tb.39.1.020
[ISO] International Organization for Standardization. 2017. Microbiology of the food chain-Horizontal method for the detection, enumeration and serotyping of Salmonella - Part 1: Detection of Salmonella spp. 6579-1:2017. [cited 2023 Jul 7]; Available from: https://www.iso.org/standard/56712.html
Li S, Zhao M, Liu J, Zhou Y, Miao Z. 2016. Prevalence and antibiotic resistance profiles of extended-spectrum β-lactamase-producing Escherichia coli isolated from healthy broilers in Shandong Province, China. J Food Prot 79(7):1169-73. DOI: https://doi.org/10.4315/0362-028X.JFP-16-025
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Monnet. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18(3):268-81. DOI: https://doi.org/10.1111/j.1469-0691.2011.03570.x
Monstein HJ, Östholm-Balkhed Å, Nilsson MV, Nilsson M, Dornbusch K, Nilsson LE. 2007. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 115(12):1400-8. DOI: https://doi.org/10.1111/j.1600-0463.2007.00722.x
Ng KCS, Rivera WL. 2015. Multiplex PCR–based serogrouping and serotyping of Salmonella enterica from tonsil and jejunum with jejunal lymph nodes of slaughtered swine in Metro Manila, Philippines. J Food Prot 78(5):873-80. DOI: https://doi.org/10.4315/0362-028X.JFP-14-342
Nguyen MN, Hoang HTT, Xavier BB, Lammens C, Le HT, Hoang NTB, Malhotra-Kumar S. 2021. Prospective One Health genetic surveillance in Vietnam identifies distinct blaCTX-M-harbouring Escherichia coli in food-chain and human-derived samples. Clin Microbiol Infect 27(10):1515.e1-1515.e8. DOI: https://doi.org/10.1016/j.cmi.2021.01.006
Paterson DL. 2000. Recommendation for treatment of severe infections caused by Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs). Clin Microbiol Infect 6(9):460-3. DOI: https://doi.org/10.1046/j.1469-0691.2000.00107.x
Pitout JDD, Hossain A, Hanson ND. 2004. Phenotypic and molecular detection of CTX-M-β-lactamases produced by Escherichia coli and Klebsiella spp. J Clin Microbiol 42(12):5715-21. DOI: https://doi.org/10.1128/JCM.42.12.5715-5721.2004
Rossolini GM, D'Andrea MM,0 Mugnaioli C. 2008. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin Microbiol Infect 14:33-41. DOI: https://doi.org/10.1111/j.1469-0691.2007.01867.x
Sales AJ, Naebi S, Nasiri R, Bannazadeh-Baghi H. 2021. The antibiotic resistance pattern and prevalence of blaTEM, blaSHV, blaCTX-M, blaPSE-1, sipB/C, and cmlA/tetR genes in Salmonella Typhimurium isolated from children with diarrhea in Tabriz, Iran. Int J Health Life Sci 7(4): e118523. DOI: 10.5812/ijhls.118523 DOI: https://doi.org/10.5812/ijhls.118523
Santos PDM, Widmer KW, Rivera WL. 2020. PCR-based detection and serovar identification of Salmonella in retail meat collected from wet markets in Metro Manila, Philippines. PLoS One 15(9):e0239457. DOI: https://doi.org/10.1371/journal.pone.0239457
Shi Q, Ye Y, Lan P, Han X, Quan J, Zhou M, Jiang Y. 2021. Prevalence and characteristics of ceftriaxone-resistant Salmonella in Children’s Hospital in Hangzhou, China. Front Microbiol 12:764-87. DOI: https://doi.org/10.3389/fmicb.2021.764787
Stanaway JD, Parisi A, Sarkar K, Blacker BF, Reiner RC, Hay SI, Crump JA. 2019. The global burden of non-typhoidal Salmonella invasive disease: A systematic analysis for the global burden of disease study 2017. Lancet Infect Dis 19(12):1312-24. DOI: https://doi.org/10.1016/j.ijid.2020.09.898
Warnes SL, Highmore CJ, Keevil CW. 2012. Horizontal transfer of antibiotic resistance genes on abiotic touch surfaces: Implications for public health. Bassler B (Editor). mBio 3(6). DOI: https://doi.org/10.1128/mBio.00489-12
Wu H, Wang Y, Wu Y, Qiao J, Li H, Zheng S, Yang B. 2015. Emergence of β-lactamases and extended-spectrum β-lactamases (ESBLs) producing Salmonella in retail raw chicken in China. Foodborne Pathog Dis 12(3):228-34. DOI: https://doi.org/10.1089/fpd.2014.1859
Copyright (c) 2024 Miles Madayag, Mr., Ms., Ms., Windell Rivera

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Authors who publish with this journal agree with the following terms:
- Authors retain copyright and grant the journal right of first publication, with the work 1 year after publication simultaneously licensed under a Creative Commons attribution-noncommerical-noderivates 4.0 International License that allows others to share, copy and redistribute the work in any medium or format, but only where the use is for non-commercial purposes and an acknowledgement of the work's authorship and initial publication in this journal is mentioned.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See The Effect of Open Access).