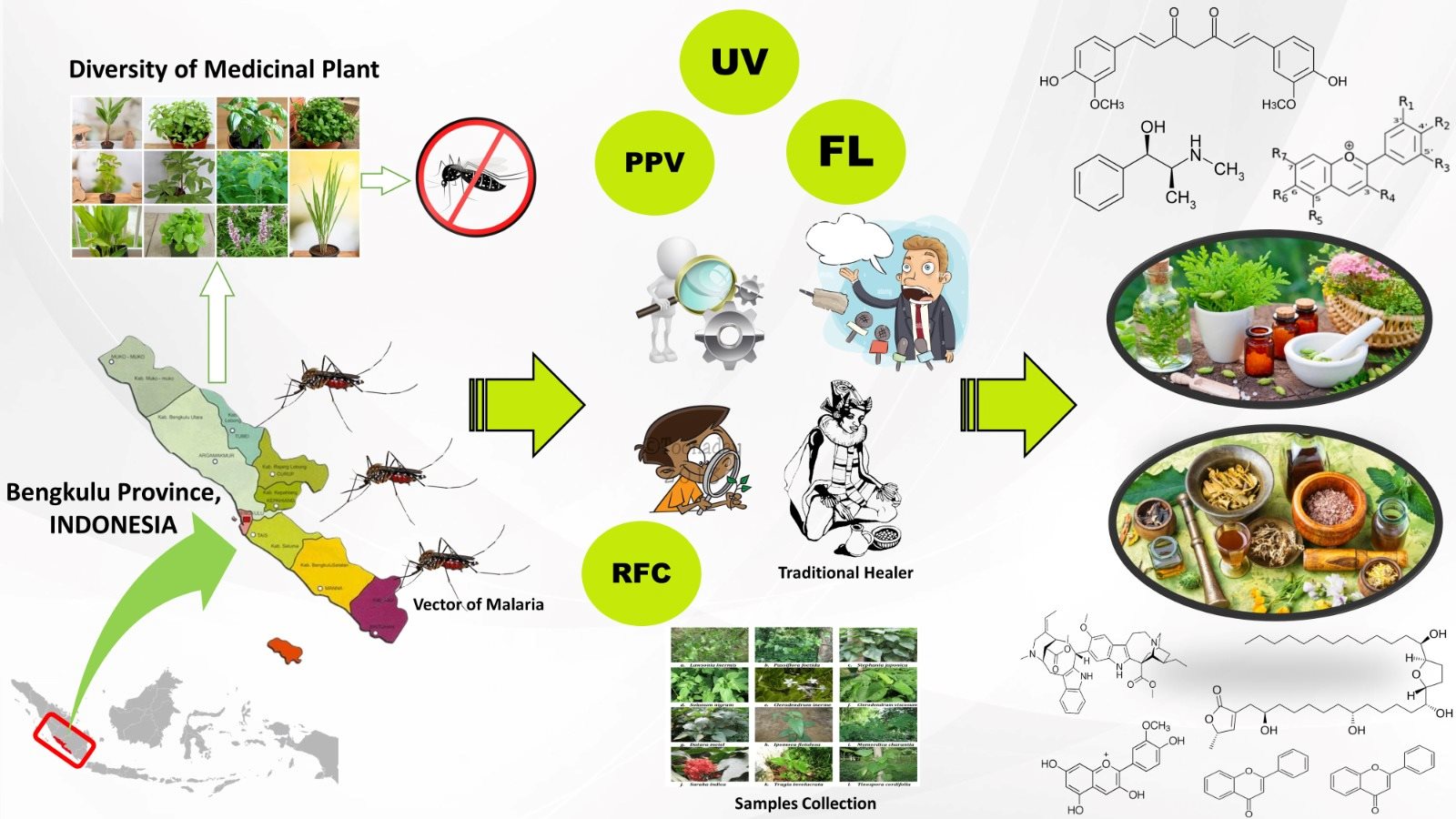
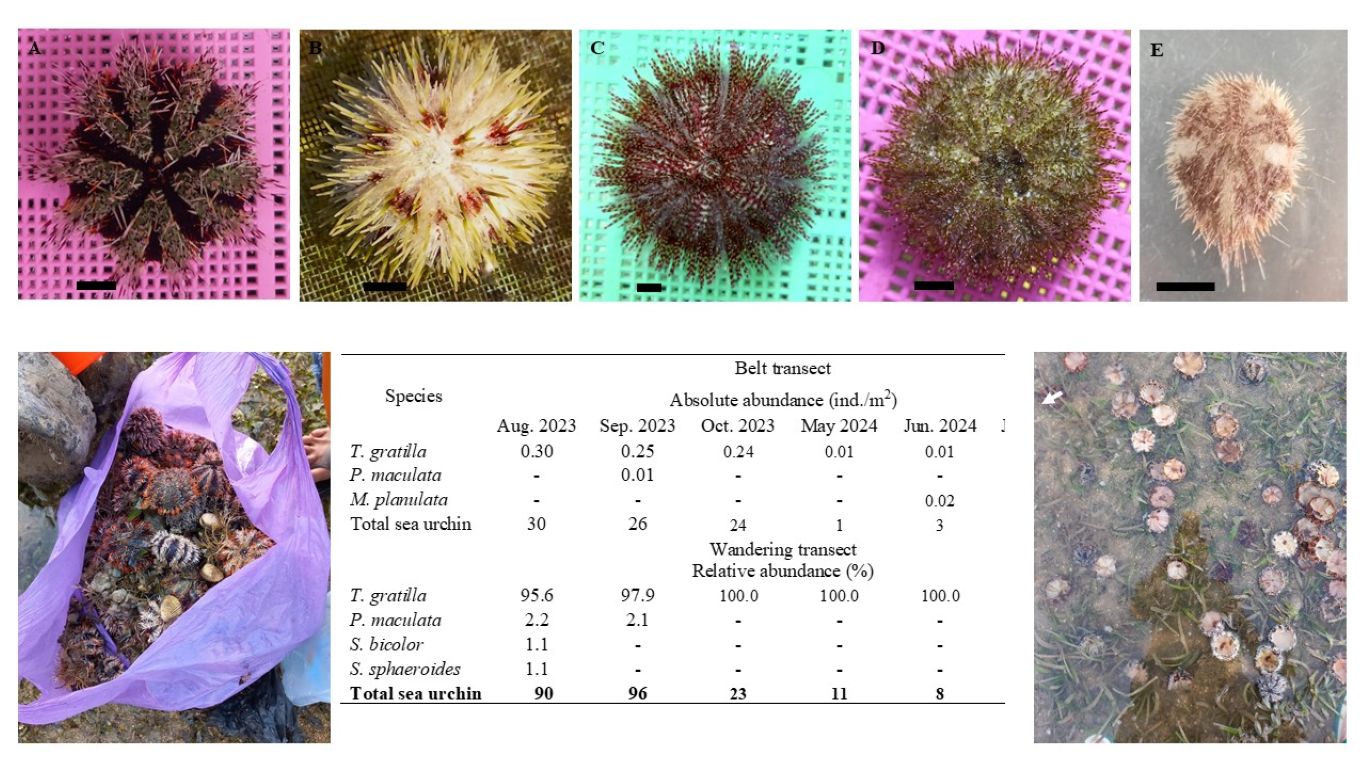
MOLECULAR DOCKING ANALYSIS OF SELECTED CURCUMA XANTHORRHIZA CONSTITUENTS AS POTENTIAL ANTICANCER DRUG
Downloads
Stress conditions will trigger the histone hyperacetylation process due to histone acetyltransferase p300/CBP (HAT PCAF) constantly transfers acetyl groups from acetyl-CoA to conserved lysine residues on histone proteins to form ε-N-acetyllysine. This can be a cause of cancer. The purpose of this study was to investigate the potential mechanisms and inhibition of PCAF HAT by chemical components of C. xanthorrhiza namely, curcumin, demethoxycurcumin, bisdemethoxycurcumin, and xanthorrizhol using in silico, the molecular docking method. Results showed that the components of C. xanthorrhiza as ligands have the capability to inhibit the binding of acetyl-CoA to histone. These results can be used to predict the inhibitory mechanisms exhibited by C. xanthorrhiza components, as competitive and noncompetitive substances. We hypothesize that C. xanthorrhiza components resemble a substrate, leading to prevention of the natural substrate (histone) to bind to the enzyme, and hence block the product formation. The smallest free Gibs energy was exhibited by curcumin on chain B and by bismethoxycurcumin on chain A, with values of -8.8 and -8.4 Kcal/mol, respectively.
Downloads
Alavijeh MS, Chisthy M, Qaiser MZ, Palmer AM. 2005. Drug metabolism and pharmacokinetics, the blood-brain barrier, and central nervous system drug discovery. NeuroRx 2:554-71. DOI: https://doi.org/10.1602/neurorx.2.4.554
Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK. 2004. Cucurmin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, repress the acetylation of histone/nonhistone proteins and histoneacetyltransferase-dependant chromatin transcription. J Biol Chem 281:40292-301. DOI: https://doi.org/10.1074/jbc.M409024200
Bimonte S, Barbecri A, Palma G, Rea D, Luciano A, D’Aluto M, Arra C, Izzo F. 2015. Dissecting the role of curcumin in tumor growth and angiogenesis in mouse model of human breast cancer. Biomed Res Int’l. ID878134. 7 pages. DOI: https://doi.org/10.1155/2015/878134
Chattopadhyay I, Biswas K, Bandyopadhyay U, Banerjee RK. 2004. Turmeric and curcumin: Biological actions and medicinal applications. Current Sci 87:44-53.
Cheah YH, Azimahtol HL, Abdullah NR. 2006. Xanthorrhizol exhibits anti-proliferative activity on MCF-7 breast cancer cells via apoptosis induction. Anticancer Res 26:4527-34.
Clements A, Rojas JR, Trievel RC, Wang L, Berger SL, Marmorstein R. 1999. Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO 18:3521-32. DOI: https://doi.org/10.1093/emboj/18.13.3521
Dekker FJ, Haisma HJ. 2009. Histone acetyltransferase as emerging drug target. Drug Discov Today 14:942-8. DOI: https://doi.org/10.1016/j.drudis.2009.06.008
Dekker FJ, Ghizzoni M, Meer NVD, Wisastra R, Haisma HJ. 2009. Inhibition of the PCAF histone acetyl transferase and cell proliferation by isothiazolones. Bioorg Med Chem 17:460-6. DOI: https://doi.org/10.1016/j.bmc.2008.12.008
Edwards MP, Price DA. 2010. Role of physicochemical properties and ligand lipophilicity efficiency in addressing drug safety risks. Annu Report Med Chem 45:381-91. DOI: https://doi.org/10.1016/S0065-7743(10)45023-X
Fan-Qi Z, Shi-Ming P, Li LLM, Zhen-Hua Z, Zhi-Yuan Z, Niu H. 2013. Structure based identification of drug-like inhibitors of p300 histone acetyltransferase. Acta Pharm Sinica 48:700-8.
Ghizzoni M, Boltjes A, Graaf CD, Haisma HJ, Dekker FJ. 2010. Improved inhibition of the histone acetyltransferase PCAF by an anacardic acid derivative. Bioorg Med Chem 18:5826-34. DOI: https://doi.org/10.1016/j.bmc.2010.06.089
Gupta S, Prasad S, Kim JH, Patchva S, Webb LJ, Priyadarsini IK, Aggarwal BB. 2011. Multitargeting by curcumin as revealed by molecular interaction studies. Nat Prod Rep 28: 1937-55. DOI: https://doi.org/10.1039/c1np00051a
Hasan AEZ, Mangunwidjaja D, Sunarti TC, Suparno O, Setiyono A. 2016. Antibreast cancer activity of nanopropolis Indonesia on induced mammary gland tumor by DMBA in virgin sprague-dawley rats. BIOTROPIA 23(1):35-41. DOI: https://doi.org/10.11598/btb.2016.23.1.473
Huang ZJ, Curtin KD, Rosbash M. 1995. PER protein interactions and temperature compensation of a circadian clock in Drosophila. Sci 267:1169-72. DOI: https://doi.org/10.1126/science.7855598
Jantan I, Saputri FC, Qaisar MN, Buang F. 2012. Correlation between chemical composition of Curcuma domestica and Curcuma xanthorrhiza and their anti-oxidant effect on human low-density lipoprotein oxidation. Evidence-Based Compl Alternative Med 438356:1-10. DOI: https://doi.org/10.1155/2012/438356
Joe B, Vijaykumar M, Lokesh BR. 2004. Biological properties of curcumin-cellular and molecular mechanisms of action. Crit Rev Food Sci Nutr 44(2):97-111. DOI: https://doi.org/10.1080/10408690490424702
Lipinski CA, Lombardo F, Segawa T, Ko D. 2001. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development setting. Adv Drug Deliv Rev 46:3-26. DOI: https://doi.org/10.1016/S0169-409X(00)00129-0
Love IM, Sekaric P, Shi D, Grossman SR, Androphy EJ. 2012. The histone acetyltransferase PCAF regulates p21 transcription through stress-induced acetylation of histone H3. Cell Cycle 11(13):2458-66. DOI: https://doi.org/10.4161/cc.20864
Mannhold R, Poda GI, Ostermann C, Tetko IV. 2007. Calculation of molecular lipophilicity: State of the art and comparison of log P methods on more than 96000 compounds. J Pharm Sci 98:861-93. DOI: https://doi.org/10.1002/jps.21494
Martz E. 2003. Introduction to bioinformatics. HAT trick: p300, Small molecule, inhibitor. Chem Biol 17:417-8. DOI: https://doi.org/10.1016/j.chembiol.2010.05.002
Monika G, Punam G, Sarbjot S, Gupta GD. 2010. An overview on molecular docking. Int J Drug Dev Res 2:219-31.
Pérez S, Tvaroška I. 2014. Carbohydrate-protein interactions: molecular modeling insights. Adv Carbohydr Chem Biochem 71:9-136. DOI: https://doi.org/10.1016/B978-0-12-800128-8.00001-7
Ropero S, Esteller M. 2007. The role of histone deacetylases (HDACs) in human cancer. Mol Oncology 1:19-25. DOI: https://doi.org/10.1016/j.molonc.2007.01.001
Sandur SK, Pandey MK, Sung B, Ahn KS, Murakami A, Sethi G, Limtrakul P, Badmaev V, Aggarwal BB. 2007. Curcumin, demethoxycurcumin, bismethoxycurcumin, tetrahydrocurcumin and termerones differentially regulate anti-inflammatory and anti-proliferative responses through a ROS-independent mechanism. Carcin 28:1765-73. DOI: https://doi.org/10.1093/carcin/bgm123
Salleh NAM, Ismail S, Halim MRA. 2016. Effects of Curcuma xanthorrhiza extracts and their constituents on phase II drug-metabolizing enzymes activity. Pharmacognosy Res 8:309-15. DOI: https://doi.org/10.4103/0974-8490.188873
Sharma NK, Jha KK, Priyanka 2010. Molecular docking: An overview. J Adv Sci Res 1:67-72.
Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95(7):927-37. DOI: https://doi.org/10.1016/S0092-8674(00)81717-1
Vinsentricia A. 2015. Kinetika inhibisi dan analisis in silico molekuler kurkumin kunyit (Curcuma longa) terhadap histon asetiltransferase. [Tesis]. Bogor (ID): Institut Pertanian Bogor.
Wapenaar H, Dekker FJ. 2016. Histone acetyltransferases: Challenges in targeting bi-substrate enzymes. Clin Epigenet 8:1-11. DOI: https://doi.org/10.1186/s13148-016-0225-2
Copyright (c) 2023 akhmad endangzainal hasan

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Authors who publish with this journal agree with the following terms:
- Authors retain copyright and grant the journal right of first publication, with the work 1 year after publication simultaneously licensed under a Creative Commons attribution-noncommerical-noderivates 4.0 International License that allows others to share, copy and redistribute the work in any medium or format, but only where the use is for non-commercial purposes and an acknowledgement of the work's authorship and initial publication in this journal is mentioned.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See The Effect of Open Access).